Happy Holidays, from my family to yours! Check back with us in early 2013 for more beautiful cells!
-Erin
December 17, 2012
Today’s image is from a paper on the development of the zebrafish eye lens. As I look at the paper’s stunning images, I’m sure that these cute little eyes are following me around my room.
The eye lens is composed of two cell types called lens epithelial cells and lens fibers. During development of the eye, lens fibers are generated from dividing lens epithelial cells, and later undergo morphogenesis. During morphogenesis, these new lens fibers elongate and then migrate toward the midline of lens, with newer lens fibers displacing and compacting older fiber layers as the entire lens grows. A recent paper investigates the relationship between morphogenesis and cell interactions with the underlying extracellular matrix (ECM). Hayes and colleagues found that fibronectin1 (Fn1), an ECM component, and integrin α5, the Fn1 cellular binding partner, are both required for lens fiber morphogenesis in developing zebrafish. Mutations in either gene cause defects in lens fiber adhesion, elongation, and packing, in turn leading to cataracts. Hayes and colleagues suggest that lens fibers migrate along an Fn1-containing substrate, which in turn activates the signaling required for lens fiber morphogenesis. In the images above, Fn1 is labeled in the developing lens of control zebrafish (top) and fn1 mutants (bottom). The control lens shows Fn1 (red) at the apical side of the lens epithelium (asterisks) and in lens fibers at the posterior (arrows), while the mutant shows reduced levels of Fn1.
 Hayes, J., Hartsock, A., Clark, B., Napier, H., Link, B., & Gross, J. (2012). Integrin 5/fibronectin1 and focal adhesion kinase are required for lens fiber morphogenesis in zebrafish Molecular Biology of the Cell, 23 (24), 4725-4738 DOI: 10.1091/mbc.E12-09-0672
Hayes, J., Hartsock, A., Clark, B., Napier, H., Link, B., & Gross, J. (2012). Integrin 5/fibronectin1 and focal adhesion kinase are required for lens fiber morphogenesis in zebrafish Molecular Biology of the Cell, 23 (24), 4725-4738 DOI: 10.1091/mbc.E12-09-0672
The eye lens is composed of two cell types called lens epithelial cells and lens fibers. During development of the eye, lens fibers are generated from dividing lens epithelial cells, and later undergo morphogenesis. During morphogenesis, these new lens fibers elongate and then migrate toward the midline of lens, with newer lens fibers displacing and compacting older fiber layers as the entire lens grows. A recent paper investigates the relationship between morphogenesis and cell interactions with the underlying extracellular matrix (ECM). Hayes and colleagues found that fibronectin1 (Fn1), an ECM component, and integrin α5, the Fn1 cellular binding partner, are both required for lens fiber morphogenesis in developing zebrafish. Mutations in either gene cause defects in lens fiber adhesion, elongation, and packing, in turn leading to cataracts. Hayes and colleagues suggest that lens fibers migrate along an Fn1-containing substrate, which in turn activates the signaling required for lens fiber morphogenesis. In the images above, Fn1 is labeled in the developing lens of control zebrafish (top) and fn1 mutants (bottom). The control lens shows Fn1 (red) at the apical side of the lens epithelium (asterisks) and in lens fibers at the posterior (arrows), while the mutant shows reduced levels of Fn1.
Labels:
cell migration,
development,
zebrafish
December 13, 2012
DNA is decorated more beautifully than a Christmas tree, but the decorations are not just for looks—they serve the important function of regulating which genes get expressed, and when. Today’s image is from a paper describing the role of a protein that affects gene expression by regulating the accessibility of some regions of the DNA.
Chromatin is the packaged DNA and associated proteins found in the nucleus. Certain regions of chromatin are packaged or modified to make the underlying genes more or less accessible to the transcription machinery that results in gene expression. A recent paper describes the identification of UpSET, a fruit fly protein that binds to promoter regions of transcriptionally active genes. The upSET gene resembles the mammalian gene MLL5, which is found in a region frequently deleted in a subset of leukemias. In the absence of UpSET, according to Rincon-Arano and colleagues, cells have increased chromatin accessibility and express genes that normally flank the regions of UpSET binding. In the images above, fruit fly polytene chromosomes (blue) show staining for UpSET (green) in gene-rich regions.
 Rincon-Arano, H., Halow, J., Delrow, J., Parkhurst, S., & Groudine, M. (2012). UpSET Recruits HDAC Complexes and Restricts Chromatin Accessibility and Acetylation at Promoter Regions Cell, 151 (6), 1214-1228 DOI: 10.1016/j.cell.2012.11.009
Rincon-Arano, H., Halow, J., Delrow, J., Parkhurst, S., & Groudine, M. (2012). UpSET Recruits HDAC Complexes and Restricts Chromatin Accessibility and Acetylation at Promoter Regions Cell, 151 (6), 1214-1228 DOI: 10.1016/j.cell.2012.11.009
Copyright ©2012 Elsevier Ltd. All rights reserved.
Chromatin is the packaged DNA and associated proteins found in the nucleus. Certain regions of chromatin are packaged or modified to make the underlying genes more or less accessible to the transcription machinery that results in gene expression. A recent paper describes the identification of UpSET, a fruit fly protein that binds to promoter regions of transcriptionally active genes. The upSET gene resembles the mammalian gene MLL5, which is found in a region frequently deleted in a subset of leukemias. In the absence of UpSET, according to Rincon-Arano and colleagues, cells have increased chromatin accessibility and express genes that normally flank the regions of UpSET binding. In the images above, fruit fly polytene chromosomes (blue) show staining for UpSET (green) in gene-rich regions.
Copyright ©2012 Elsevier Ltd. All rights reserved.
Labels:
DNA,
Drosophila,
genes
December 10, 2012
For some types of cells, notably polarized cells, the localization of a protein can be regulated through mRNAs. mRNAs are transcribed from DNA, and then later translated into the proteins that function throughout the cell. By transporting mRNAs to specific regions, the cell in turn can have localized levels of proteins. A recent paper shows the specific localization of an mRNA encoding the signaling molecule MKK7 to neuronal growth cones, which are dynamic extension of a developing axon searching for its final target. According to Feltrin and colleagues, this localization of MKK7 mRNA may result in localized levels of MKK7 protein. MKK7 mRNA localization modulates JNK signaling, which in turn regulates microtubule bundling during neuronal outgrowth. In the images above, cells with reduced levels of MKK7 mRNA (bottom) have curled and bent microtubules (red in merged, black in right panels), compared to control cells (top).
Labels:
microtubules,
mRNA,
neurons
December 6, 2012
When developmental biology and cell biology combine, I get absolutely giddy thinking about the microscopy advances and fabulous images involved. Imaging a single cell is difficult enough, but the microscopy challenges facing biologists who study developing organisms is enough to make some run the other way. Today’s images are from a paper describing the role of prostaglandins in actin remodeling in the fruit fly egg chamber.
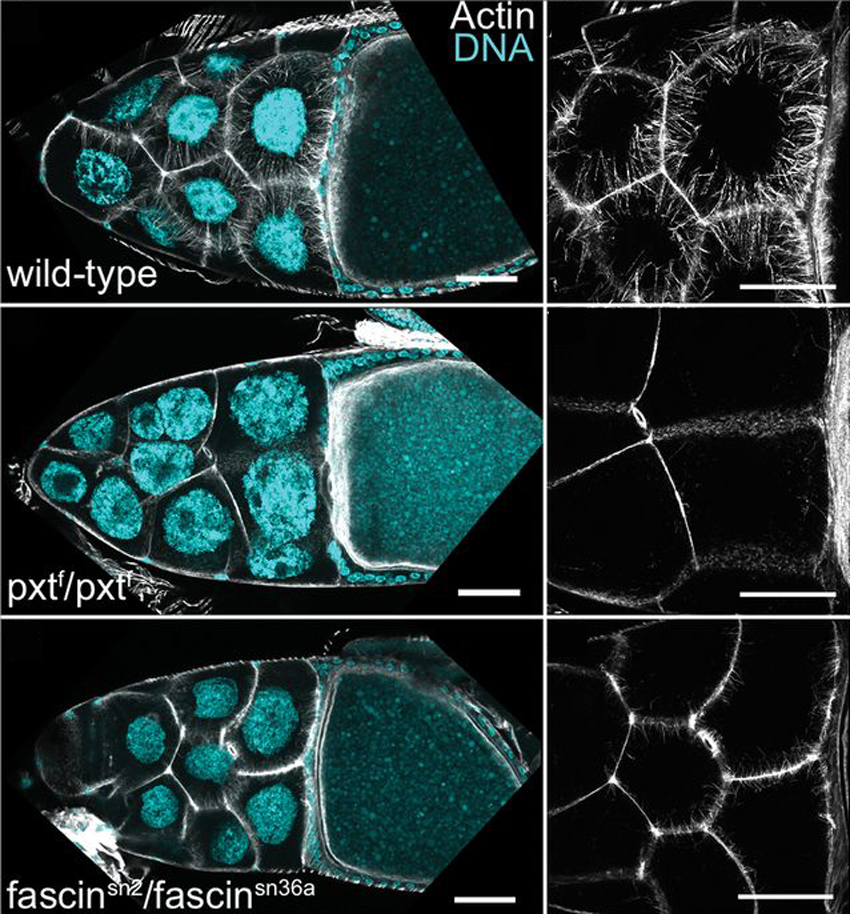
Prostaglandins (PGs) are lipid signaling molecules that regulate a wide range of processes, such as inflammation, pain, and hormone regulation. One target of PGs is the actin cytoskeleton, but how PGs affect actin filament polymerization and dynamics is not well understood. A recent paper uses the fruit fly egg chamber as a great model of how PGs regulate actin. In the fruit fly egg chamber, many nurse cells surround the oocyte (developing egg). These nurse cells are squeezed in order to dump all of their cytoplasmic contents through ring canals and into the growing egg. During this process, the nuclei of nurse cells are held in place by bundles of actin filaments to ensure that the nuclei don’t plug up the ring canals and block the transfer of the cytoplasm. PGs are an important part of this process, and they interact with an actin-bundling protein called Fascin, according to a recent paper by Groen and colleagues. Inhibition of either Fascin or Pxt, an enzyme required for PG production, results in the failure of actin bundle formation and nurse cell dumping. The loss of Fascin enhances the effects of Pxt reduction, and the overexpression of fascin suppresses the defects seen in flies with Pxt loss. Groen and colleagues show that PGs regulate Fascin, which modulates actin cytoskeleton rearrangements during nurse cell dumping. In the images above, control egg chambers (top) show parallel actin bundles (white) within nurse cells, which extend from the cell membranes to the nuclei (blue). pxt and fascin mutants (middle, bottom), however, contain little to no actin bundles in nurse cells.
 Groen, C., Spracklen, A., Fagan, T., & Tootle, T. (2012). Drosophila Fascin is a novel downstream target of prostaglandin signaling during actin remodeling Molecular Biology of the Cell, 23 (23), 4567-4578 DOI: 10.1091/mbc.E12-05-0417
Groen, C., Spracklen, A., Fagan, T., & Tootle, T. (2012). Drosophila Fascin is a novel downstream target of prostaglandin signaling during actin remodeling Molecular Biology of the Cell, 23 (23), 4567-4578 DOI: 10.1091/mbc.E12-05-0417
Prostaglandins (PGs) are lipid signaling molecules that regulate a wide range of processes, such as inflammation, pain, and hormone regulation. One target of PGs is the actin cytoskeleton, but how PGs affect actin filament polymerization and dynamics is not well understood. A recent paper uses the fruit fly egg chamber as a great model of how PGs regulate actin. In the fruit fly egg chamber, many nurse cells surround the oocyte (developing egg). These nurse cells are squeezed in order to dump all of their cytoplasmic contents through ring canals and into the growing egg. During this process, the nuclei of nurse cells are held in place by bundles of actin filaments to ensure that the nuclei don’t plug up the ring canals and block the transfer of the cytoplasm. PGs are an important part of this process, and they interact with an actin-bundling protein called Fascin, according to a recent paper by Groen and colleagues. Inhibition of either Fascin or Pxt, an enzyme required for PG production, results in the failure of actin bundle formation and nurse cell dumping. The loss of Fascin enhances the effects of Pxt reduction, and the overexpression of fascin suppresses the defects seen in flies with Pxt loss. Groen and colleagues show that PGs regulate Fascin, which modulates actin cytoskeleton rearrangements during nurse cell dumping. In the images above, control egg chambers (top) show parallel actin bundles (white) within nurse cells, which extend from the cell membranes to the nuclei (blue). pxt and fascin mutants (middle, bottom), however, contain little to no actin bundles in nurse cells.
Labels:
actin,
development,
Drosophila
December 3, 2012
Macrophages are our little Pac-Man cells that gobble up the nasty pathogens that we may come into contact with. They’re importance is undeniable, and the images of macrophages doing their thing are always fascinating to me. Today’s image is from a paper describing the role of the protein Bcl10 in the process.
Phagocytosis is the process during which a cell engulfs cellular debris or pathogens. Macrophages employ phagocytosis to rid the body of infectious agents, cellular debris in the lung, and necrotic tissue throughout the body. Phagocytosis requires regulation of actin polymerization and dynamics for the extension and then closure of a phagocytic cup that engulfs the target material. A recent paper describes the role of the NF-κB signaling protein Bcl10 in actin and membrane remodeling in human macrophages. Marion and colleagues found that without Bcl10, phagocytosis begins but then stops with accumulated actin. Bcl10 regulates actin polymerization in cups, which allows the vesicle exocytosis that causes membrane extension of the phagocytic cup. In the images above, macrophages were incubated with red blood cells (red) to track phagocytosis 5 minutes (left column) and 10 minutes (right column) after the start of the assay. Control macrophages (top) are able to phagocytose red blood cells after a few minutes into the assay (arrows point to membrane deformations indicative of phagocytosed particles). Cells lacking Bcl10 (bottom), however, are not able to complete engulfment of the red blood cells, and have shorter and disorganized membrane extensions compared with control cells.
 Marion, S., Mazzolini, J., Herit, F., Bourdoncle, P., Kambou-Pene, N., Hailfinger, S., Sachse, M., Ruland, J., Benmerah, A., Echard, A., Thome, M., & Niedergang, F. (2012). The NF-κB Signaling Protein Bcl10 Regulates Actin Dynamics by Controlling AP1 and OCRL-Bearing Vesicles Developmental Cell, 23 (5), 954-967 DOI: 10.1016/j.devcel.2012.09.021
Marion, S., Mazzolini, J., Herit, F., Bourdoncle, P., Kambou-Pene, N., Hailfinger, S., Sachse, M., Ruland, J., Benmerah, A., Echard, A., Thome, M., & Niedergang, F. (2012). The NF-κB Signaling Protein Bcl10 Regulates Actin Dynamics by Controlling AP1 and OCRL-Bearing Vesicles Developmental Cell, 23 (5), 954-967 DOI: 10.1016/j.devcel.2012.09.021
Copyright ©2012 Elsevier Ltd. All rights reserved.
Phagocytosis is the process during which a cell engulfs cellular debris or pathogens. Macrophages employ phagocytosis to rid the body of infectious agents, cellular debris in the lung, and necrotic tissue throughout the body. Phagocytosis requires regulation of actin polymerization and dynamics for the extension and then closure of a phagocytic cup that engulfs the target material. A recent paper describes the role of the NF-κB signaling protein Bcl10 in actin and membrane remodeling in human macrophages. Marion and colleagues found that without Bcl10, phagocytosis begins but then stops with accumulated actin. Bcl10 regulates actin polymerization in cups, which allows the vesicle exocytosis that causes membrane extension of the phagocytic cup. In the images above, macrophages were incubated with red blood cells (red) to track phagocytosis 5 minutes (left column) and 10 minutes (right column) after the start of the assay. Control macrophages (top) are able to phagocytose red blood cells after a few minutes into the assay (arrows point to membrane deformations indicative of phagocytosed particles). Cells lacking Bcl10 (bottom), however, are not able to complete engulfment of the red blood cells, and have shorter and disorganized membrane extensions compared with control cells.
Copyright ©2012 Elsevier Ltd. All rights reserved.
Labels:
phagocytosis
November 30, 2012
I didn’t pay enough attention to primary cilia in my earlier years, and that is one of life’s big regrets (well, regret is a strong word). They are very fascinating little sensory organelles, and the thought of primary cilia carrying the weight of neuron migration on their little basal body shoulders is impressive. Check out today’s image, from a paper showing the role of primary cilia in brain development.
Neurons are frequently born far from their final home in the brain, and this migration is key to healthy nervous system development and function. A recent paper shows the importance of primary cilia in the migration of interneurons (neurons that connect one neuron to another) in the cerebral cortex. Primary cilia are microtubule-based sensory organelles that project out of a cell’s membrane. Higginbotham and colleagues imaged migrating interneurons in the developing cerebral cortex and found a correlation between primary cilia dynamics and interneuron mobility. This process requires the ciliary protein Arl13b, a GTPase in the Arf/Arl family. Arl13b ensures correct localization and movement of guidance cue receptors in primary cilia. In the images above, interneurons (green chamber in cartoon, green cells in images) migrate along tracks toward a signal secreted by dorsal cortical cells (blue chamber in cartoon). The migration of Arl13b mutant interneurons (right panel) was drastically reduced when compared to control interneurons (left, same scale).
 Higginbotham, H., Eom, T., Mariani, L., Bachleda, A., Hirt, J., Gukassyan, V., Cusack, C., Lai, C., Caspary, T., & Anton, E. (2012). Arl13b in Primary Cilia Regulates the Migration and Placement of Interneurons in the Developing Cerebral Cortex Developmental Cell, 23 (5), 925-938 DOI: 10.1016/j.devcel.2012.09.019
Higginbotham, H., Eom, T., Mariani, L., Bachleda, A., Hirt, J., Gukassyan, V., Cusack, C., Lai, C., Caspary, T., & Anton, E. (2012). Arl13b in Primary Cilia Regulates the Migration and Placement of Interneurons in the Developing Cerebral Cortex Developmental Cell, 23 (5), 925-938 DOI: 10.1016/j.devcel.2012.09.019
Copyright ©2012 Elsevier Ltd. All rights reserved.
Neurons are frequently born far from their final home in the brain, and this migration is key to healthy nervous system development and function. A recent paper shows the importance of primary cilia in the migration of interneurons (neurons that connect one neuron to another) in the cerebral cortex. Primary cilia are microtubule-based sensory organelles that project out of a cell’s membrane. Higginbotham and colleagues imaged migrating interneurons in the developing cerebral cortex and found a correlation between primary cilia dynamics and interneuron mobility. This process requires the ciliary protein Arl13b, a GTPase in the Arf/Arl family. Arl13b ensures correct localization and movement of guidance cue receptors in primary cilia. In the images above, interneurons (green chamber in cartoon, green cells in images) migrate along tracks toward a signal secreted by dorsal cortical cells (blue chamber in cartoon). The migration of Arl13b mutant interneurons (right panel) was drastically reduced when compared to control interneurons (left, same scale).
Copyright ©2012 Elsevier Ltd. All rights reserved.
Labels:
cell migration,
cilia,
neurons
November 27, 2012
We have a ton of neurons. And each of those neurons has many dendrites. And each dendrite has countless little dendritic spines. Thinking about how complex one single neuron is in order to receive a signal from another cell gives me an identity crisis. What if we are all just little dendritic spines on our universe’s neuron?! Was that stupid stampede for Walmart’s Black Friday sale worth it for those folks? Was my self-restraint when faced with leftover pumpkin pie worth it? Is any of it worth it?!
Dendritic spines are small actin-rich protrusions on a neuron’s dendrite, the structure that receives information from other neurons. The morphology and density of the dendritic spines can be regulated by neurotransmitters, actin dynamics, and actin-regulating proteins. The neuron-specific actin regulator cortactin-binding protein 2 (CTTNBP2) regulates the formation and maintenance of dendritic spines, and is even associated with autism spectrum disorder. A recent paper investigates the role of a CTTNBP2 homologue, CTTNBP2NL (CTTNBP2 N-terminal-like protein). Chen and colleagues found that while CTTNBP2 expression is found in the brain, CTTNBP2NL is not. In addition, CTTNBP2NL does not appear to play a role in dendritic spine formation. Although both CTTNBP2 and CTTNBP2NL associate with cortactin, a well-studied actin regulator, CTTNBP2 is associated with the cell’s cortex while CTTNBP2NL is found on actin stress fibers. In addition, Chen and colleagues found a link between CTTNBP2 and the protein phosphatase 2A (PP2A) complex, specifically with CTTNBP2 targeting the PP2A complex to dendritic spines. In the images above, cells show labels for cortactin (red) and actin fibers (blue). CTTNBP2NL (green, top) associates with stress fibers (arrows), while CTTNBP2 (green, bottom) is distributed around the cortex (arrowheads).
Small World
Every year, Nikon hosts the Small World competition, which pits stunning microscopy images against one another for oooohs and aaaahs from around the world. The images are breathtaking, for both scientists and lay-folk alike
So, to kick off a brief Thanksgiving break, I wanted to send you all to the 2012 winners of the Small World competition, announced a few weeks ago. Click here for the Small World winners and honorable mentions...and lose an afternoon to image browsing, microscope jealousy, and pride in the amazing advances within the microscopy field.
Happy Thanksgiving, everyone! Thanks for reading HighMag!
So, to kick off a brief Thanksgiving break, I wanted to send you all to the 2012 winners of the Small World competition, announced a few weeks ago. Click here for the Small World winners and honorable mentions...and lose an afternoon to image browsing, microscope jealousy, and pride in the amazing advances within the microscopy field.
Happy Thanksgiving, everyone! Thanks for reading HighMag!
Labels:
fun stuff
November 12, 2012
When you were still developing, your brain was an overachiever just like that straight-A class president with perfect teeth and a canned food drive. Your brain overproduced neurons, then later paired down the neuron population to fine-tune development and function. Today’s image is from a paper that describes this process and the regulation behind it.
Interneurons are neurons that make connections with other neurons, and are found throughout our bodies. In our brain, our cortical neurons are produced far away from their final destination in the fully mature brain. It has been suggested that these neurons are overproduced, and then migrate to the cortex where the excess neurons are eliminated. A recent paper shows this process occurring in developing mice. Southwell and colleagues showed this developmental cell death occurring within the developing mouse brain, within laboratory cultures, and within cortical neurons transplanted into a developing mouse brain. Their results suggest that the cell death is triggered cell-autonomously (from within the cell) or triggered due to competition between other interneurons for survival signals. The image above shows interneuron precursor cells cultured on a plate of cortical feeder layers containing neurons (green), astrocytes (red), and oligodendrocytes (white). About 30% of the cortical interneurons cultured on these feeder layers later underwent cell death.
 Southwell, D., Paredes, M., Galvao, R., Jones, D., Froemke, R., Sebe, J., Alfaro-Cervello, C., Tang, Y., Garcia-Verdugo, J., Rubenstein, J., Baraban, S., & Alvarez-Buylla, A. (2012). Intrinsically determined cell death of developing cortical interneurons Nature, 491 (7422), 109-113 DOI: 10.1038/nature11523
Southwell, D., Paredes, M., Galvao, R., Jones, D., Froemke, R., Sebe, J., Alfaro-Cervello, C., Tang, Y., Garcia-Verdugo, J., Rubenstein, J., Baraban, S., & Alvarez-Buylla, A. (2012). Intrinsically determined cell death of developing cortical interneurons Nature, 491 (7422), 109-113 DOI: 10.1038/nature11523
Adapted by permission from Macmillan Publishers Ltd, copyright ©2012
Interneurons are neurons that make connections with other neurons, and are found throughout our bodies. In our brain, our cortical neurons are produced far away from their final destination in the fully mature brain. It has been suggested that these neurons are overproduced, and then migrate to the cortex where the excess neurons are eliminated. A recent paper shows this process occurring in developing mice. Southwell and colleagues showed this developmental cell death occurring within the developing mouse brain, within laboratory cultures, and within cortical neurons transplanted into a developing mouse brain. Their results suggest that the cell death is triggered cell-autonomously (from within the cell) or triggered due to competition between other interneurons for survival signals. The image above shows interneuron precursor cells cultured on a plate of cortical feeder layers containing neurons (green), astrocytes (red), and oligodendrocytes (white). About 30% of the cortical interneurons cultured on these feeder layers later underwent cell death.
Adapted by permission from Macmillan Publishers Ltd, copyright ©2012
Labels:
development,
neurons
November 9, 2012
Size really does matter, folks! Ask any scientist about the potential uses for nanoparticles, and you’ll quickly agree that these tiny little suckers deserve a spot at your dinner table, complete with cute nano-plates and nano-forks. Today’s image is from a paper describing a clever use of nanoparticles to systematically monitor how cells respond to force.
A cell encounters force from its entire environment. A cell responds to mechanical force by regulating signals, development, and migration, among other things. Biologists have been trying to understand how cells respond to force for years, and clever techniques have included manipulation by optical tweezers, micropipettes, and atomic force microscopy. These techniques, however, have been limited by their ability to monitor one cell at a time. A recent paper describes a technique in which many cells respond to uniform forces on the cortex. In this study, Tseng and colleagues plated cells on micropatterned magnetic substrates. Magnetic nanoparticles within the cells were then manipulated by a magnetic field in order to apply a uniform mechanical force on the cell cortex. By monitoring large numbers of individual cells under these forces, Tseng and colleagues developed a way for biologists to have higher control and accuracy in understanding how force affects cells. The images above show representative images of cells under varying amounts of force. The dose of nanoparticles (blue) in each cell (increasing from bottom to top) and strength of the magnetic field (increasing from left to right) both affect the response from cells. A higher nanoparticle dose and magnetic field gradient caused cells to produce more filopodial membrane extensions from cells (actin is in green) from the region where the force was applied.
 Tseng, P., Judy, J., & Di Carlo, D. (2012). Magnetic nanoparticle–mediated massively parallel mechanical modulation of single-cell behavior Nature Methods, 9 (11), 1113-1119 DOI: 10.1038/nmeth.2210
Tseng, P., Judy, J., & Di Carlo, D. (2012). Magnetic nanoparticle–mediated massively parallel mechanical modulation of single-cell behavior Nature Methods, 9 (11), 1113-1119 DOI: 10.1038/nmeth.2210
Adapted by permission from Macmillan Publishers Ltd, copyright ©2012
A cell encounters force from its entire environment. A cell responds to mechanical force by regulating signals, development, and migration, among other things. Biologists have been trying to understand how cells respond to force for years, and clever techniques have included manipulation by optical tweezers, micropipettes, and atomic force microscopy. These techniques, however, have been limited by their ability to monitor one cell at a time. A recent paper describes a technique in which many cells respond to uniform forces on the cortex. In this study, Tseng and colleagues plated cells on micropatterned magnetic substrates. Magnetic nanoparticles within the cells were then manipulated by a magnetic field in order to apply a uniform mechanical force on the cell cortex. By monitoring large numbers of individual cells under these forces, Tseng and colleagues developed a way for biologists to have higher control and accuracy in understanding how force affects cells. The images above show representative images of cells under varying amounts of force. The dose of nanoparticles (blue) in each cell (increasing from bottom to top) and strength of the magnetic field (increasing from left to right) both affect the response from cells. A higher nanoparticle dose and magnetic field gradient caused cells to produce more filopodial membrane extensions from cells (actin is in green) from the region where the force was applied.
Adapted by permission from Macmillan Publishers Ltd, copyright ©2012
Labels:
techniques
November 6, 2012
It is Election Day here in the US, which means that some of us need stress relief until the results are tallied. So, maybe you can imagine the graceful membrane dynamics of a cell and let their lava-lamp-like groove lull you into a happy place. Today’s image is from a paper describing the regulation of caveola biogenesis.
Caveolae are small invaginations on a cell’s plasma membrane that play important roles in cell signaling and endocytosis (the uptake of material into a cell). Caveolae depend on proteins called caveolins, but the details of caveola biogenesis are not completely understood. A recent paper describes results showing the importance of phosphorylation of Caveolin-1 (Cav1) in the formation of caveolae. Phosphorylation is the addition of a phosphate group to a protein, which functions as a molecular switch to change the protein’s activity. Joshi and colleagues found that the phosphorylation of Cav1 induces a feedback loop that links together mechanical stress on the cell, caveola biogenesis, and focal adhesion regulation at the cell’s membrane. These results place Cav1 on a list of critical proteins that help a cell respond to mechanical stress. In the images above, mutations of Cav1 that mimic phosphorylation (Cav1Y14R and Cav1Y14D) cause the formation of more caveolae and caveolae clusters (arrows and asterisks) than control cells (Cav1WT).
 Joshi, B., Bastiani, M., Strugnell, S., Boscher, C., Parton, R., & Nabi, I. (2012). Phosphocaveolin-1 is a mechanotransducer that induces caveola biogenesis via Egr1 transcriptional regulation originally published in the Journal of Cell Biology, 199 (3), 425-435 DOI: 10.1083/jcb.201207089
Joshi, B., Bastiani, M., Strugnell, S., Boscher, C., Parton, R., & Nabi, I. (2012). Phosphocaveolin-1 is a mechanotransducer that induces caveola biogenesis via Egr1 transcriptional regulation originally published in the Journal of Cell Biology, 199 (3), 425-435 DOI: 10.1083/jcb.201207089
Caveolae are small invaginations on a cell’s plasma membrane that play important roles in cell signaling and endocytosis (the uptake of material into a cell). Caveolae depend on proteins called caveolins, but the details of caveola biogenesis are not completely understood. A recent paper describes results showing the importance of phosphorylation of Caveolin-1 (Cav1) in the formation of caveolae. Phosphorylation is the addition of a phosphate group to a protein, which functions as a molecular switch to change the protein’s activity. Joshi and colleagues found that the phosphorylation of Cav1 induces a feedback loop that links together mechanical stress on the cell, caveola biogenesis, and focal adhesion regulation at the cell’s membrane. These results place Cav1 on a list of critical proteins that help a cell respond to mechanical stress. In the images above, mutations of Cav1 that mimic phosphorylation (Cav1Y14R and Cav1Y14D) cause the formation of more caveolae and caveolae clusters (arrows and asterisks) than control cells (Cav1WT).
Labels:
membranes,
phosphorylation
November 2, 2012
Despite being a scientist, sci-fi/fantasy is just not my cup of tea. Sometimes, though, I am positive that a scientific name is really some Klingon starship or Game of Throne character. Ever since I learned about the nodes of Ranvier in high school biology, I have been sure that they’re really from some fantasy world. Today’s image is from a paper that doesn’t really dispel my confusion...the concept of measuring and understanding high nerve conduction velocity in teeny tiny axons is other-worldly.
Myelin is a material that forms a layer around the axon of a neuron. Schwann cells wrap around axons and produce these myelin sheaths, which are spaced between gaps called the nodes of Ranvier. The main purpose of myelin is to allow nerve impulses to move very quickly along the axon, but the relationship between nerve conduction velocity and the distance between myelin sheaths was unclear. Recently, Wu and colleagues measured conduction velocity in mice with Schwann cells carrying a mutation that prevented elongation of Schwann cells. In these cells with short Schwann cells, and in turn short distances between nodes of Ranvier, conduction velocity dropped and motor function of the mice was impaired. As these mice developed and the internodal distance increased, nerve conduction velocity and motor function recovered. Wu and colleagues suggest that the high conduction speed reached by increasing internodal distance reaches a “flat maximum.” Above, cross-sections of nerves in mice at 3 (top) or 24 (bottom) weeks old show some differences in myelin between normal mice (left column) and mice with a Schwann cell elongation mutation (right column). 24-week old mutants show some myelin folds and some structures indicative of demyelination and remyelination (arrowheads, bottom right).
 Wu, L., Williams, A., Delaney, A., Sherman, D., & Brophy, P. (2012). Increasing Internodal Distance in Myelinated Nerves Accelerates Nerve Conduction to a Flat Maximum Current Biology, 22 (20), 1957-1961 DOI: 10.1016/j.cub.2012.08.025
Wu, L., Williams, A., Delaney, A., Sherman, D., & Brophy, P. (2012). Increasing Internodal Distance in Myelinated Nerves Accelerates Nerve Conduction to a Flat Maximum Current Biology, 22 (20), 1957-1961 DOI: 10.1016/j.cub.2012.08.025
Copyright ©2012 Elsevier Ltd. All rights reserved.
Myelin is a material that forms a layer around the axon of a neuron. Schwann cells wrap around axons and produce these myelin sheaths, which are spaced between gaps called the nodes of Ranvier. The main purpose of myelin is to allow nerve impulses to move very quickly along the axon, but the relationship between nerve conduction velocity and the distance between myelin sheaths was unclear. Recently, Wu and colleagues measured conduction velocity in mice with Schwann cells carrying a mutation that prevented elongation of Schwann cells. In these cells with short Schwann cells, and in turn short distances between nodes of Ranvier, conduction velocity dropped and motor function of the mice was impaired. As these mice developed and the internodal distance increased, nerve conduction velocity and motor function recovered. Wu and colleagues suggest that the high conduction speed reached by increasing internodal distance reaches a “flat maximum.” Above, cross-sections of nerves in mice at 3 (top) or 24 (bottom) weeks old show some differences in myelin between normal mice (left column) and mice with a Schwann cell elongation mutation (right column). 24-week old mutants show some myelin folds and some structures indicative of demyelination and remyelination (arrowheads, bottom right).
Copyright ©2012 Elsevier Ltd. All rights reserved.
Labels:
neurons
October 30, 2012
I’ve posted a lot of images from papers on human disease models lately, and that’s a good thing. Despite what you may hear from politicians mocking research using fruit flies, worms, or tiny fish, the power of model systems to study a human problem is undeniable to anyone with half a clue. All science on the continuum from basic research all the way to clinical trials is valuable…so thank your favorite scientist today! Today’s image is from a zebrafish paper that identifies a new pathway that may be a good therapy target for muscular dystrophies.
Muscular dystrophies are fairly common diseases that result in the weakening of the protein complexes that connect muscles to their underlying environment, called the extracellular matrix (ECM). The degeneration of muscle-ECM attachment eventually leads to progressive loss of muscle movement and locomotion for the patient. A recent paper identifies a new pathway in muscle-ECM attachment using zebrafish as a model. Goody and colleagues found that biosynthesis of the small molecule NAD+ can reverse muscle degeneration in certain types of dystrophies in zebrafish and even improve swimming. This pathway improves the organization of laminin, an important ECM protein, suggesting the function of an additional laminin receptor complex and pathway from those already studied. Goody and colleagues suggest that this new pathway may serve as a focal point for new muscular dystrophy therapies. In the images above, normal ECM basement membrane tissue adhered normally to both normal muscle cells (blue) and cells with muscle degeneration mutations (red), even after the stress of swimming. This experiment shows the importance of a healthy ECM in restoring function for dystrophic muscles.
 Goody, M., Kelly, M., Reynolds, C., Khalil, A., Crawford, B., & Henry, C. (2012). NAD+ Biosynthesis Ameliorates a Zebrafish Model of Muscular Dystrophy PLoS Biology, 10 (10) DOI: 10.1371/journal.pbio.1001409
Goody, M., Kelly, M., Reynolds, C., Khalil, A., Crawford, B., & Henry, C. (2012). NAD+ Biosynthesis Ameliorates a Zebrafish Model of Muscular Dystrophy PLoS Biology, 10 (10) DOI: 10.1371/journal.pbio.1001409
Muscular dystrophies are fairly common diseases that result in the weakening of the protein complexes that connect muscles to their underlying environment, called the extracellular matrix (ECM). The degeneration of muscle-ECM attachment eventually leads to progressive loss of muscle movement and locomotion for the patient. A recent paper identifies a new pathway in muscle-ECM attachment using zebrafish as a model. Goody and colleagues found that biosynthesis of the small molecule NAD+ can reverse muscle degeneration in certain types of dystrophies in zebrafish and even improve swimming. This pathway improves the organization of laminin, an important ECM protein, suggesting the function of an additional laminin receptor complex and pathway from those already studied. Goody and colleagues suggest that this new pathway may serve as a focal point for new muscular dystrophy therapies. In the images above, normal ECM basement membrane tissue adhered normally to both normal muscle cells (blue) and cells with muscle degeneration mutations (red), even after the stress of swimming. This experiment shows the importance of a healthy ECM in restoring function for dystrophic muscles.
Labels:
disease,
extracellular,
muscle
October 26, 2012
Animal models of human diseases allow biologists to move quickly on understanding the physiology behind a disease and make suggestions on developing therapies. Today’s image is from a paper showing the importance of using multiple animal models for the same human disease.
The genetic disorder Usher syndrome (USH) causes hearing and visual impairment. Mice have provided biologists with a fantastic model of hearing impairment in USH patients, specifically those with the most severe form of the syndrome, USH1. The five USH1 proteins (myosin VIIa, harmonin, cadherin-23, protocadherin-15, sans) all play a role in the development of the hair bundle, the structure that drives the conversion of sounds into electrical responses in the ear. Mice with USH1 mutations do not, however, experience the same visual degeneration as human USH1 patients. A recent study finds differences in USH1 protein localization in the photoreceptors of different animal models. Sahly and colleagues found that the photoreceptors of macaques (monkeys) show localization of all USH1 proteins at membrane interfaces, while photoreceptors in mice lack any USH1 network proteins. Sahly and colleagues suggest that the USH1 network found in humans and other primates forms an adhesion belt around the basolateral region of photoreceptors, and this structure may be the key to understanding the visual impairment experienced by USH1 patients. In the images above of macaque retina photoreceptors, the USH1 network protein protocadherin-15 (green) is localized at the junction between the inner (IS) and outer segments (OS) of rods and cones.
 Sahly, I., Dufour, E., Schietroma, C., Michel, V., Bahloul, A., Perfettini, I., Pepermans, E., Estivalet, A., Carette, D., Aghaie, A., Ebermann, I., Lelli, A., Iribarne, M., Hardelin, J., Weil, D., Sahel, J., El-Amraoui, A., & Petit, C. (2012). Localization of Usher 1 proteins to the photoreceptor calyceal processes, which are absent from mice originally published in the Journal of Cell Biology, 199 (2), 381-399 DOI: 10.1083/jcb.201202012
Sahly, I., Dufour, E., Schietroma, C., Michel, V., Bahloul, A., Perfettini, I., Pepermans, E., Estivalet, A., Carette, D., Aghaie, A., Ebermann, I., Lelli, A., Iribarne, M., Hardelin, J., Weil, D., Sahel, J., El-Amraoui, A., & Petit, C. (2012). Localization of Usher 1 proteins to the photoreceptor calyceal processes, which are absent from mice originally published in the Journal of Cell Biology, 199 (2), 381-399 DOI: 10.1083/jcb.201202012
The genetic disorder Usher syndrome (USH) causes hearing and visual impairment. Mice have provided biologists with a fantastic model of hearing impairment in USH patients, specifically those with the most severe form of the syndrome, USH1. The five USH1 proteins (myosin VIIa, harmonin, cadherin-23, protocadherin-15, sans) all play a role in the development of the hair bundle, the structure that drives the conversion of sounds into electrical responses in the ear. Mice with USH1 mutations do not, however, experience the same visual degeneration as human USH1 patients. A recent study finds differences in USH1 protein localization in the photoreceptors of different animal models. Sahly and colleagues found that the photoreceptors of macaques (monkeys) show localization of all USH1 proteins at membrane interfaces, while photoreceptors in mice lack any USH1 network proteins. Sahly and colleagues suggest that the USH1 network found in humans and other primates forms an adhesion belt around the basolateral region of photoreceptors, and this structure may be the key to understanding the visual impairment experienced by USH1 patients. In the images above of macaque retina photoreceptors, the USH1 network protein protocadherin-15 (green) is localized at the junction between the inner (IS) and outer segments (OS) of rods and cones.
Labels:
disease
October 23, 2012
Understanding how a cell works normally is hard enough for biologists. Understanding how a cancerous cell works is exponentially harder—there are different stages of tumorigenesis and countless different types of cancer and countless different environments within the body. Today’s image is from a study that takes a systematic approach to understanding the interactions between cancerous cells and their environment.
The progression of tumor cells to metastatic cancer cells correlates with poor prognoses for cancer patients. The steps that drive cancer cells to spread (metastasis) are not well understood, but may be effective targets for chemotherapies. For example, tumor cells lose adhesion to their underlying extracellular matrix (ECM) prior to spreading to other regions of the body. Understanding this loss of cell-ECM adhesion may guide the development of new therapies. A recent paper describes the systematic analysis of cell-ECM adhesion in tumor cells by using robotically spotted arrays of 768 paired combinations of ECM molecules. Reticker-Flynn and colleagues monitored the adhesion profiles of lung cancer cell lines at different stages of cancer progression on these arrays of ECM molecules, and found ECM-cell interactions that may be successful therapeutic targets. The images above show an array of spotted ECM protein combinations (visible through immuno- and fluorescent-labeling, top), and examples of cells adhered to the ECM spots (bottom images).
 Reticker-Flynn, N., Malta, D., Winslow, M., Lamar, J., Xu, M., Underhill, G., Hynes, R., Jacks, T., & Bhatia, S. (2012). A combinatorial extracellular matrix platform identifies cell-extracellular matrix interactions that correlate with metastasis Nature Communications, 3 DOI: 10.1038/ncomms2128
Reticker-Flynn, N., Malta, D., Winslow, M., Lamar, J., Xu, M., Underhill, G., Hynes, R., Jacks, T., & Bhatia, S. (2012). A combinatorial extracellular matrix platform identifies cell-extracellular matrix interactions that correlate with metastasis Nature Communications, 3 DOI: 10.1038/ncomms2128
Adapted by permission from Macmillan Publishers Ltd, copyright ©2012
The progression of tumor cells to metastatic cancer cells correlates with poor prognoses for cancer patients. The steps that drive cancer cells to spread (metastasis) are not well understood, but may be effective targets for chemotherapies. For example, tumor cells lose adhesion to their underlying extracellular matrix (ECM) prior to spreading to other regions of the body. Understanding this loss of cell-ECM adhesion may guide the development of new therapies. A recent paper describes the systematic analysis of cell-ECM adhesion in tumor cells by using robotically spotted arrays of 768 paired combinations of ECM molecules. Reticker-Flynn and colleagues monitored the adhesion profiles of lung cancer cell lines at different stages of cancer progression on these arrays of ECM molecules, and found ECM-cell interactions that may be successful therapeutic targets. The images above show an array of spotted ECM protein combinations (visible through immuno- and fluorescent-labeling, top), and examples of cells adhered to the ECM spots (bottom images).
Adapted by permission from Macmillan Publishers Ltd, copyright ©2012
Labels:
adhesion,
cancer,
disease,
extracellular
October 19, 2012
The next time you see a fruit fly buzzing around your kitchen, take a beat before you smack it with a swatter and remind yourself of the amazing discoveries due to organisms like the (not so) lowly fruit fly. Maybe you’ll offer a thank-you glass of grape juice instead and show the intruder back outside. Today’s image is from a paper describing a fly model of a serious human disease, and serves as a great example of the power in a model organism.
Spinal muscular atrophy (SMA) is a heritable disease that results in infant mortality due motor neuron dysfunction and rapid degeneration of muscle. SMA is caused by the depletion of the SMN (survival motor neuron) protein. A recent paper describes the use of fruit flies in studying SMA, and shows that flies lacking SMN have reduced muscle size and defective motor neuron neurotransmission similar to SMA patients. Imlach and colleagues found that replenishing SMN levels in motor neurons and muscles did not reverse the defects of SMN depletion, yet increasing SMN levels in partner cells (proprioceptive neurons and interneurons) can reverse the defects. These results suggest that SMN depletion primarily affects the sensory-motor network, with secondary effects seen in the motor circuit. In addition, Imlach and colleagues found that increasing motor neural circuit excitability, either genetically or with drugs, could relieve SMN-depletion defects. Using this fly model of SMA, these results suggest that SMA patients may improve by enhancing motor neural network activity. The images above show the differences in muscle size in wild-type (left) and SMN-depleted (right) flies.
 Imlach WL, Beck ES, Choi BJ, Lotti F, Pellizzoni L, & McCabe BD (2012). SMN Is Required for Sensory-Motor Circuit Function in Drosophila. Cell, 151 (2), 427-39 PMID: 23063130
Imlach WL, Beck ES, Choi BJ, Lotti F, Pellizzoni L, & McCabe BD (2012). SMN Is Required for Sensory-Motor Circuit Function in Drosophila. Cell, 151 (2), 427-39 PMID: 23063130
Copyright ©2012 Elsevier Ltd. All rights reserved.
Spinal muscular atrophy (SMA) is a heritable disease that results in infant mortality due motor neuron dysfunction and rapid degeneration of muscle. SMA is caused by the depletion of the SMN (survival motor neuron) protein. A recent paper describes the use of fruit flies in studying SMA, and shows that flies lacking SMN have reduced muscle size and defective motor neuron neurotransmission similar to SMA patients. Imlach and colleagues found that replenishing SMN levels in motor neurons and muscles did not reverse the defects of SMN depletion, yet increasing SMN levels in partner cells (proprioceptive neurons and interneurons) can reverse the defects. These results suggest that SMN depletion primarily affects the sensory-motor network, with secondary effects seen in the motor circuit. In addition, Imlach and colleagues found that increasing motor neural circuit excitability, either genetically or with drugs, could relieve SMN-depletion defects. Using this fly model of SMA, these results suggest that SMA patients may improve by enhancing motor neural network activity. The images above show the differences in muscle size in wild-type (left) and SMN-depleted (right) flies.
Copyright ©2012 Elsevier Ltd. All rights reserved.
Labels:
disease,
Drosophila,
muscle,
neurons
October 16, 2012
Rho GTPases are actin regulators that switch between active and inactive states. Their activation state is regulated by proteins (GEFs) that drive the GTPases into their active state, and others (GAPs) that drive them into their inactive state. RhoGDIs (Rho guanine nucleotide dissociation inhibitors) provide an additional level of control over Rho GTPase activity by sequestering Rho GTPases in inactive complexes. The selective dissociation of individual Rho GTPases from these complexes provides the cell with a context-specific response based on the actin-based structures required. A recent paper describes the function of a protein called diacylglycerol kinase ζ (DGKζ) in the release of two Rho GTPases, Rac1 and RhoA, from RhoGDI complexes. Rac1 regulates membrane ruffling and lamellipodia formation, while RhoA regulates stress fiber and focal adhesion formation. Ard and colleagues found that DGKζ-deficient cells showed signs of faulty RhoA signaling, as seen in the images above. Actin stress fibers (left column, green in merged) and focal adhesions (middle column, red in merged) appear normal in wild-type cells (top). In DGKζ-null cells (bottom row), actin stress fibers appeared condensed (arrow) and less organized, while focal adhesion distribution was impaired.
Labels:
actin,
focal adhesions
October 11, 2012
When in a crowded elevator, mall, or football stadium, I panic knowing that I’d be the first person trampled and/or eaten in an emergency. Crowded places give me the willies, but when proteins are over-crowded, interesting things happen. Today’s image is from a paper showing what can happen at the membrane when proteins are over-crowded.
Membrane bending is an essential part of many cellular events, including endocytosis and filopodia formation. It has been suggested that membrane curvature can stem from two mechanisms—the use of curved proteins to form a curved scaffold for the membrane, or the insertion of wedge-shaped hydrophobic helices into the membrane. A recent paper shows a third mechanism that can drive membrane curvature. Stachowiak and colleagues found that protein-protein crowding drives membrane bending. Protein coverage at 20% is sufficient to drive curvature, by creating lateral pressure of membrane-bound proteins colliding. Interestingly, even proteins unrelated to membrane curvature can induce curvature when overcrowded (GFP, for example). The cartoons and images above show vesicles that contain low (left) or high (right) levels of protein binding at specific domains at the membrane. High levels of protein caused membrane curvature, as seen as long lipid tubules.
 Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, & Hayden CC (2012). Membrane bending by protein-protein crowding. Nature cell biology, 14 (9), 944-9 PMID: 22902598
Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, & Hayden CC (2012). Membrane bending by protein-protein crowding. Nature cell biology, 14 (9), 944-9 PMID: 22902598
Adapted by permission from Macmillan Publishers Ltd, copyright ©2012
Membrane bending is an essential part of many cellular events, including endocytosis and filopodia formation. It has been suggested that membrane curvature can stem from two mechanisms—the use of curved proteins to form a curved scaffold for the membrane, or the insertion of wedge-shaped hydrophobic helices into the membrane. A recent paper shows a third mechanism that can drive membrane curvature. Stachowiak and colleagues found that protein-protein crowding drives membrane bending. Protein coverage at 20% is sufficient to drive curvature, by creating lateral pressure of membrane-bound proteins colliding. Interestingly, even proteins unrelated to membrane curvature can induce curvature when overcrowded (GFP, for example). The cartoons and images above show vesicles that contain low (left) or high (right) levels of protein binding at specific domains at the membrane. High levels of protein caused membrane curvature, as seen as long lipid tubules.
Adapted by permission from Macmillan Publishers Ltd, copyright ©2012
Labels:
membranes
October 8, 2012
If you are a developmental biologist, there is a high probability that you study Wnt. The Wnt signaling pathway is employed throughout cell and developmental biology in processes ranging from spindle positioning to stem cell fate decisions. Today’s image is from a paper showing how Wnt can be secreted from cells.
Wnt signaling functions by relaying a signal from the cell’s surface to the nucleus, where gene expression is regulated. Active Wnt proteins are secreted out of a cell to induce tissue patterning during development (among many other things), and can travel over a distance of several cells. In trying to understand Wnt secretion, a recent paper describes results showing that active Wnt signals can be secreted from exosomes. Exosomes are vesicles that cells use to secrete various materials into the extracellular space. Gross and colleagues found that Wnt signals are secreted on exosomes in both developing fruit flies and human cells. Wnt signals are trafficked through various endosomal compartments and then to exosomes, with the help of the trafficking protein Ykt6 (an R-SNARE). In the top row of images above, Wnt (left column, red in merged) appears colocalized with an exosomal protein (CD63, green in merged) in developing fly wing discs. Wnt is also colocalized with a marker for multivesicular bodies (LAMP-1, green in merged), vesicles from which exosomes originate. Insets show higher magnification views of the vesicles.
 Gross JC, Chaudhary V, Bartscherer K, & Boutros M (2012). Active Wnt proteins are secreted on exosomes. Nature cell biology, 14 (10), 1036-45 PMID: 22983114
Gross JC, Chaudhary V, Bartscherer K, & Boutros M (2012). Active Wnt proteins are secreted on exosomes. Nature cell biology, 14 (10), 1036-45 PMID: 22983114
Adapted by permission from Macmillan Publishers Ltd, copyright ©2012
Wnt signaling functions by relaying a signal from the cell’s surface to the nucleus, where gene expression is regulated. Active Wnt proteins are secreted out of a cell to induce tissue patterning during development (among many other things), and can travel over a distance of several cells. In trying to understand Wnt secretion, a recent paper describes results showing that active Wnt signals can be secreted from exosomes. Exosomes are vesicles that cells use to secrete various materials into the extracellular space. Gross and colleagues found that Wnt signals are secreted on exosomes in both developing fruit flies and human cells. Wnt signals are trafficked through various endosomal compartments and then to exosomes, with the help of the trafficking protein Ykt6 (an R-SNARE). In the top row of images above, Wnt (left column, red in merged) appears colocalized with an exosomal protein (CD63, green in merged) in developing fly wing discs. Wnt is also colocalized with a marker for multivesicular bodies (LAMP-1, green in merged), vesicles from which exosomes originate. Insets show higher magnification views of the vesicles.
Adapted by permission from Macmillan Publishers Ltd, copyright ©2012
Labels:
development,
Drosophila,
exocytosis,
signaling,
Wnt
Subscribe to:
Posts (Atom)