Happy Holidays, from my family to yours! Check back with us in early 2013 for more beautiful cells!
-Erin
December 17, 2012
Today’s image is from a paper on the development of the zebrafish eye lens. As I look at the paper’s stunning images, I’m sure that these cute little eyes are following me around my room.
The eye lens is composed of two cell types called lens epithelial cells and lens fibers. During development of the eye, lens fibers are generated from dividing lens epithelial cells, and later undergo morphogenesis. During morphogenesis, these new lens fibers elongate and then migrate toward the midline of lens, with newer lens fibers displacing and compacting older fiber layers as the entire lens grows. A recent paper investigates the relationship between morphogenesis and cell interactions with the underlying extracellular matrix (ECM). Hayes and colleagues found that fibronectin1 (Fn1), an ECM component, and integrin α5, the Fn1 cellular binding partner, are both required for lens fiber morphogenesis in developing zebrafish. Mutations in either gene cause defects in lens fiber adhesion, elongation, and packing, in turn leading to cataracts. Hayes and colleagues suggest that lens fibers migrate along an Fn1-containing substrate, which in turn activates the signaling required for lens fiber morphogenesis. In the images above, Fn1 is labeled in the developing lens of control zebrafish (top) and fn1 mutants (bottom). The control lens shows Fn1 (red) at the apical side of the lens epithelium (asterisks) and in lens fibers at the posterior (arrows), while the mutant shows reduced levels of Fn1.
 Hayes, J., Hartsock, A., Clark, B., Napier, H., Link, B., & Gross, J. (2012). Integrin 5/fibronectin1 and focal adhesion kinase are required for lens fiber morphogenesis in zebrafish Molecular Biology of the Cell, 23 (24), 4725-4738 DOI: 10.1091/mbc.E12-09-0672
Hayes, J., Hartsock, A., Clark, B., Napier, H., Link, B., & Gross, J. (2012). Integrin 5/fibronectin1 and focal adhesion kinase are required for lens fiber morphogenesis in zebrafish Molecular Biology of the Cell, 23 (24), 4725-4738 DOI: 10.1091/mbc.E12-09-0672
The eye lens is composed of two cell types called lens epithelial cells and lens fibers. During development of the eye, lens fibers are generated from dividing lens epithelial cells, and later undergo morphogenesis. During morphogenesis, these new lens fibers elongate and then migrate toward the midline of lens, with newer lens fibers displacing and compacting older fiber layers as the entire lens grows. A recent paper investigates the relationship between morphogenesis and cell interactions with the underlying extracellular matrix (ECM). Hayes and colleagues found that fibronectin1 (Fn1), an ECM component, and integrin α5, the Fn1 cellular binding partner, are both required for lens fiber morphogenesis in developing zebrafish. Mutations in either gene cause defects in lens fiber adhesion, elongation, and packing, in turn leading to cataracts. Hayes and colleagues suggest that lens fibers migrate along an Fn1-containing substrate, which in turn activates the signaling required for lens fiber morphogenesis. In the images above, Fn1 is labeled in the developing lens of control zebrafish (top) and fn1 mutants (bottom). The control lens shows Fn1 (red) at the apical side of the lens epithelium (asterisks) and in lens fibers at the posterior (arrows), while the mutant shows reduced levels of Fn1.
Labels:
cell migration,
development,
zebrafish
December 13, 2012
DNA is decorated more beautifully than a Christmas tree, but the decorations are not just for looks—they serve the important function of regulating which genes get expressed, and when. Today’s image is from a paper describing the role of a protein that affects gene expression by regulating the accessibility of some regions of the DNA.
Chromatin is the packaged DNA and associated proteins found in the nucleus. Certain regions of chromatin are packaged or modified to make the underlying genes more or less accessible to the transcription machinery that results in gene expression. A recent paper describes the identification of UpSET, a fruit fly protein that binds to promoter regions of transcriptionally active genes. The upSET gene resembles the mammalian gene MLL5, which is found in a region frequently deleted in a subset of leukemias. In the absence of UpSET, according to Rincon-Arano and colleagues, cells have increased chromatin accessibility and express genes that normally flank the regions of UpSET binding. In the images above, fruit fly polytene chromosomes (blue) show staining for UpSET (green) in gene-rich regions.
 Rincon-Arano, H., Halow, J., Delrow, J., Parkhurst, S., & Groudine, M. (2012). UpSET Recruits HDAC Complexes and Restricts Chromatin Accessibility and Acetylation at Promoter Regions Cell, 151 (6), 1214-1228 DOI: 10.1016/j.cell.2012.11.009
Rincon-Arano, H., Halow, J., Delrow, J., Parkhurst, S., & Groudine, M. (2012). UpSET Recruits HDAC Complexes and Restricts Chromatin Accessibility and Acetylation at Promoter Regions Cell, 151 (6), 1214-1228 DOI: 10.1016/j.cell.2012.11.009
Copyright ©2012 Elsevier Ltd. All rights reserved.
Chromatin is the packaged DNA and associated proteins found in the nucleus. Certain regions of chromatin are packaged or modified to make the underlying genes more or less accessible to the transcription machinery that results in gene expression. A recent paper describes the identification of UpSET, a fruit fly protein that binds to promoter regions of transcriptionally active genes. The upSET gene resembles the mammalian gene MLL5, which is found in a region frequently deleted in a subset of leukemias. In the absence of UpSET, according to Rincon-Arano and colleagues, cells have increased chromatin accessibility and express genes that normally flank the regions of UpSET binding. In the images above, fruit fly polytene chromosomes (blue) show staining for UpSET (green) in gene-rich regions.
Copyright ©2012 Elsevier Ltd. All rights reserved.
Labels:
DNA,
Drosophila,
genes
December 10, 2012
For some types of cells, notably polarized cells, the localization of a protein can be regulated through mRNAs. mRNAs are transcribed from DNA, and then later translated into the proteins that function throughout the cell. By transporting mRNAs to specific regions, the cell in turn can have localized levels of proteins. A recent paper shows the specific localization of an mRNA encoding the signaling molecule MKK7 to neuronal growth cones, which are dynamic extension of a developing axon searching for its final target. According to Feltrin and colleagues, this localization of MKK7 mRNA may result in localized levels of MKK7 protein. MKK7 mRNA localization modulates JNK signaling, which in turn regulates microtubule bundling during neuronal outgrowth. In the images above, cells with reduced levels of MKK7 mRNA (bottom) have curled and bent microtubules (red in merged, black in right panels), compared to control cells (top).
Labels:
microtubules,
mRNA,
neurons
December 6, 2012
When developmental biology and cell biology combine, I get absolutely giddy thinking about the microscopy advances and fabulous images involved. Imaging a single cell is difficult enough, but the microscopy challenges facing biologists who study developing organisms is enough to make some run the other way. Today’s images are from a paper describing the role of prostaglandins in actin remodeling in the fruit fly egg chamber.
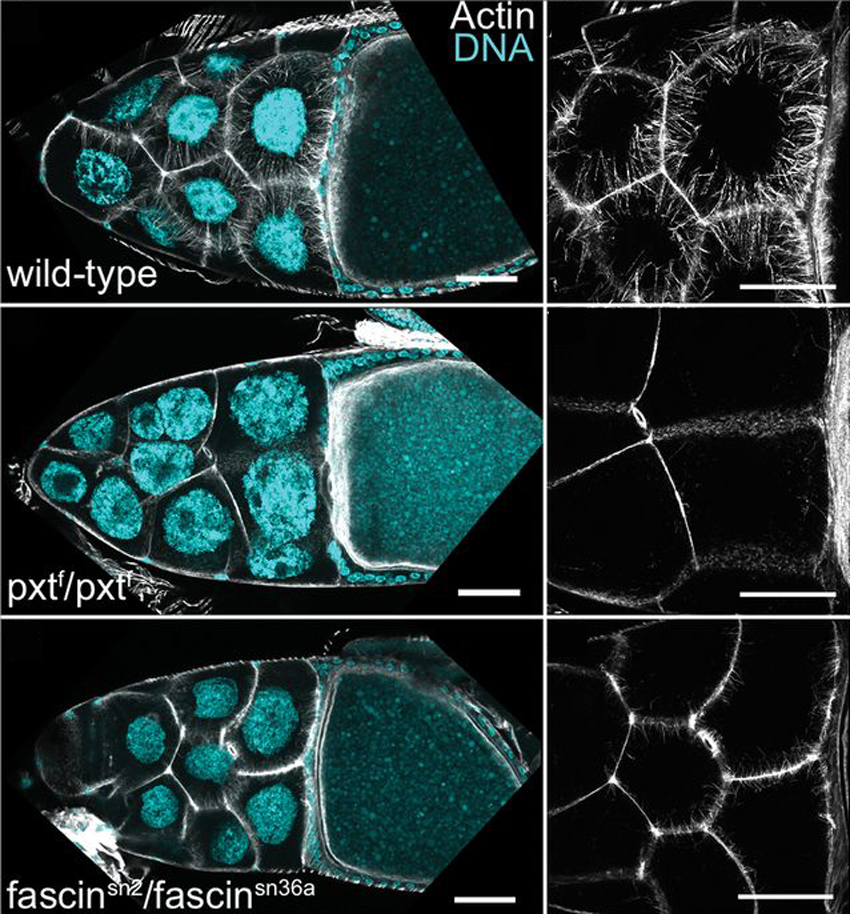
Prostaglandins (PGs) are lipid signaling molecules that regulate a wide range of processes, such as inflammation, pain, and hormone regulation. One target of PGs is the actin cytoskeleton, but how PGs affect actin filament polymerization and dynamics is not well understood. A recent paper uses the fruit fly egg chamber as a great model of how PGs regulate actin. In the fruit fly egg chamber, many nurse cells surround the oocyte (developing egg). These nurse cells are squeezed in order to dump all of their cytoplasmic contents through ring canals and into the growing egg. During this process, the nuclei of nurse cells are held in place by bundles of actin filaments to ensure that the nuclei don’t plug up the ring canals and block the transfer of the cytoplasm. PGs are an important part of this process, and they interact with an actin-bundling protein called Fascin, according to a recent paper by Groen and colleagues. Inhibition of either Fascin or Pxt, an enzyme required for PG production, results in the failure of actin bundle formation and nurse cell dumping. The loss of Fascin enhances the effects of Pxt reduction, and the overexpression of fascin suppresses the defects seen in flies with Pxt loss. Groen and colleagues show that PGs regulate Fascin, which modulates actin cytoskeleton rearrangements during nurse cell dumping. In the images above, control egg chambers (top) show parallel actin bundles (white) within nurse cells, which extend from the cell membranes to the nuclei (blue). pxt and fascin mutants (middle, bottom), however, contain little to no actin bundles in nurse cells.
 Groen, C., Spracklen, A., Fagan, T., & Tootle, T. (2012). Drosophila Fascin is a novel downstream target of prostaglandin signaling during actin remodeling Molecular Biology of the Cell, 23 (23), 4567-4578 DOI: 10.1091/mbc.E12-05-0417
Groen, C., Spracklen, A., Fagan, T., & Tootle, T. (2012). Drosophila Fascin is a novel downstream target of prostaglandin signaling during actin remodeling Molecular Biology of the Cell, 23 (23), 4567-4578 DOI: 10.1091/mbc.E12-05-0417
Prostaglandins (PGs) are lipid signaling molecules that regulate a wide range of processes, such as inflammation, pain, and hormone regulation. One target of PGs is the actin cytoskeleton, but how PGs affect actin filament polymerization and dynamics is not well understood. A recent paper uses the fruit fly egg chamber as a great model of how PGs regulate actin. In the fruit fly egg chamber, many nurse cells surround the oocyte (developing egg). These nurse cells are squeezed in order to dump all of their cytoplasmic contents through ring canals and into the growing egg. During this process, the nuclei of nurse cells are held in place by bundles of actin filaments to ensure that the nuclei don’t plug up the ring canals and block the transfer of the cytoplasm. PGs are an important part of this process, and they interact with an actin-bundling protein called Fascin, according to a recent paper by Groen and colleagues. Inhibition of either Fascin or Pxt, an enzyme required for PG production, results in the failure of actin bundle formation and nurse cell dumping. The loss of Fascin enhances the effects of Pxt reduction, and the overexpression of fascin suppresses the defects seen in flies with Pxt loss. Groen and colleagues show that PGs regulate Fascin, which modulates actin cytoskeleton rearrangements during nurse cell dumping. In the images above, control egg chambers (top) show parallel actin bundles (white) within nurse cells, which extend from the cell membranes to the nuclei (blue). pxt and fascin mutants (middle, bottom), however, contain little to no actin bundles in nurse cells.
Labels:
actin,
development,
Drosophila
December 3, 2012
Macrophages are our little Pac-Man cells that gobble up the nasty pathogens that we may come into contact with. They’re importance is undeniable, and the images of macrophages doing their thing are always fascinating to me. Today’s image is from a paper describing the role of the protein Bcl10 in the process.
Phagocytosis is the process during which a cell engulfs cellular debris or pathogens. Macrophages employ phagocytosis to rid the body of infectious agents, cellular debris in the lung, and necrotic tissue throughout the body. Phagocytosis requires regulation of actin polymerization and dynamics for the extension and then closure of a phagocytic cup that engulfs the target material. A recent paper describes the role of the NF-κB signaling protein Bcl10 in actin and membrane remodeling in human macrophages. Marion and colleagues found that without Bcl10, phagocytosis begins but then stops with accumulated actin. Bcl10 regulates actin polymerization in cups, which allows the vesicle exocytosis that causes membrane extension of the phagocytic cup. In the images above, macrophages were incubated with red blood cells (red) to track phagocytosis 5 minutes (left column) and 10 minutes (right column) after the start of the assay. Control macrophages (top) are able to phagocytose red blood cells after a few minutes into the assay (arrows point to membrane deformations indicative of phagocytosed particles). Cells lacking Bcl10 (bottom), however, are not able to complete engulfment of the red blood cells, and have shorter and disorganized membrane extensions compared with control cells.
 Marion, S., Mazzolini, J., Herit, F., Bourdoncle, P., Kambou-Pene, N., Hailfinger, S., Sachse, M., Ruland, J., Benmerah, A., Echard, A., Thome, M., & Niedergang, F. (2012). The NF-κB Signaling Protein Bcl10 Regulates Actin Dynamics by Controlling AP1 and OCRL-Bearing Vesicles Developmental Cell, 23 (5), 954-967 DOI: 10.1016/j.devcel.2012.09.021
Marion, S., Mazzolini, J., Herit, F., Bourdoncle, P., Kambou-Pene, N., Hailfinger, S., Sachse, M., Ruland, J., Benmerah, A., Echard, A., Thome, M., & Niedergang, F. (2012). The NF-κB Signaling Protein Bcl10 Regulates Actin Dynamics by Controlling AP1 and OCRL-Bearing Vesicles Developmental Cell, 23 (5), 954-967 DOI: 10.1016/j.devcel.2012.09.021
Copyright ©2012 Elsevier Ltd. All rights reserved.
Phagocytosis is the process during which a cell engulfs cellular debris or pathogens. Macrophages employ phagocytosis to rid the body of infectious agents, cellular debris in the lung, and necrotic tissue throughout the body. Phagocytosis requires regulation of actin polymerization and dynamics for the extension and then closure of a phagocytic cup that engulfs the target material. A recent paper describes the role of the NF-κB signaling protein Bcl10 in actin and membrane remodeling in human macrophages. Marion and colleagues found that without Bcl10, phagocytosis begins but then stops with accumulated actin. Bcl10 regulates actin polymerization in cups, which allows the vesicle exocytosis that causes membrane extension of the phagocytic cup. In the images above, macrophages were incubated with red blood cells (red) to track phagocytosis 5 minutes (left column) and 10 minutes (right column) after the start of the assay. Control macrophages (top) are able to phagocytose red blood cells after a few minutes into the assay (arrows point to membrane deformations indicative of phagocytosed particles). Cells lacking Bcl10 (bottom), however, are not able to complete engulfment of the red blood cells, and have shorter and disorganized membrane extensions compared with control cells.
Copyright ©2012 Elsevier Ltd. All rights reserved.
Labels:
phagocytosis
Subscribe to:
Comments (Atom)